Trajectory Generation for Automated Molecule Manipulation
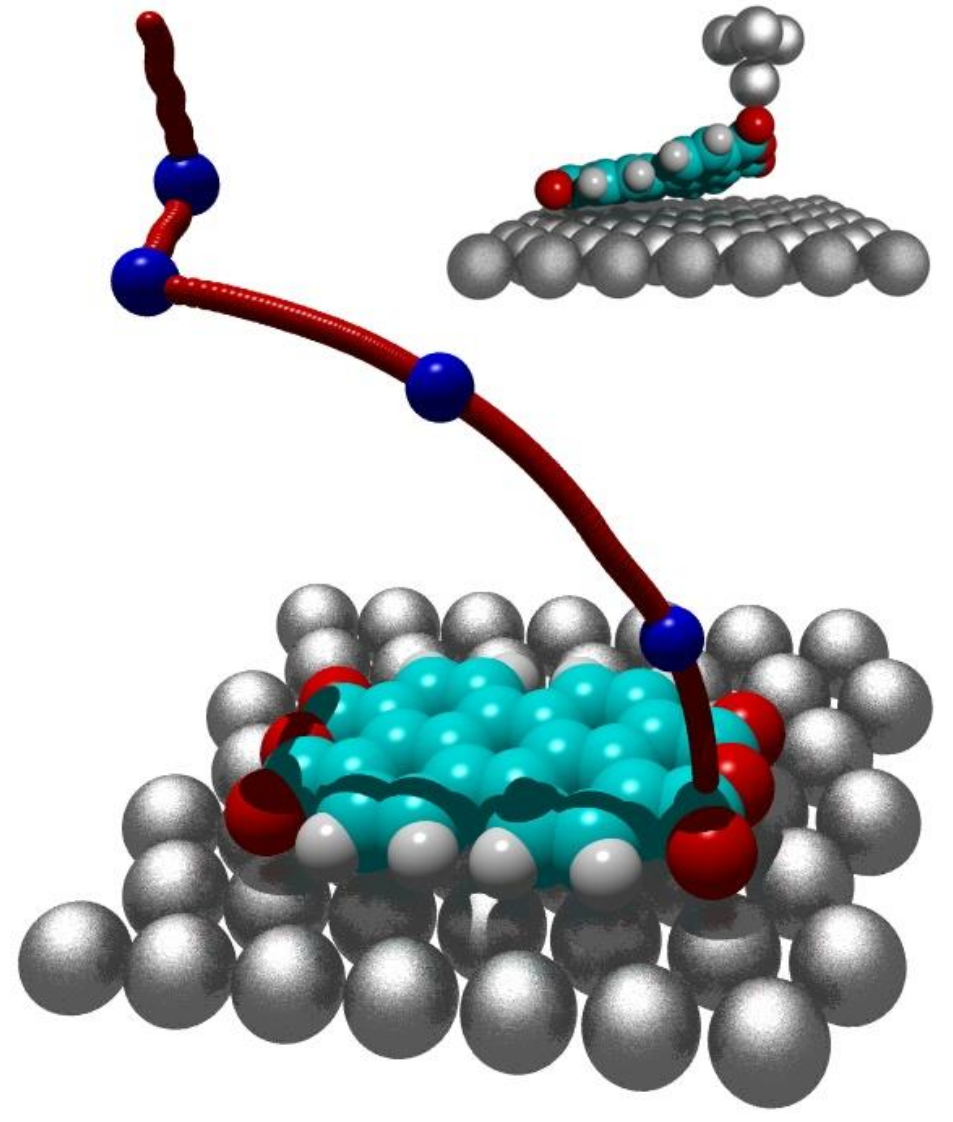
A major goal of nanotechnology is to arrange atoms or molecules one by one the way we want them. Within the scope of a collaboration with the Peter Grünberg Institut of the Jülich Research Center, we investigate how the atomic force microscope (AFM) can be used to manipulate single molecules.
This Bachelor's thesis is intended to automate this challenging task. For that purpose, we employ a simulation model that captures the necessary effects on the atomic scale to automatically generate a lifting trajectory that can be then applied to the AFM. The student has to familiarize with and extend the existing simulation model, conduct a literature research on trajectory generation, and finally choose and apply suitable approaches to the task of lifting a molecule.
The successful completion of this thesis provides a crucial ingredient towards one of the fundamental goals in nanotechnology.
Topic Area:
Helpful/Required Prerequisites:
Experience with: Matlab, C++
Language: German or English
Project Start:
Estimated time requirements:
Implementation: 40%
Validation: 40%