Research
The main focus of our research activities...
JAK-Sys
|
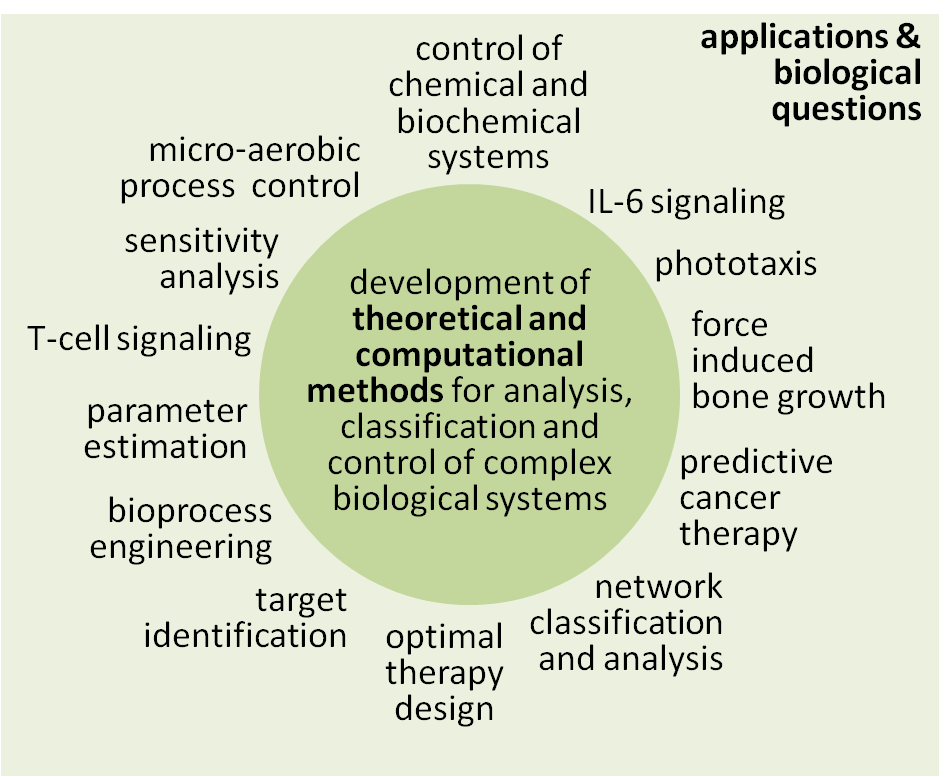
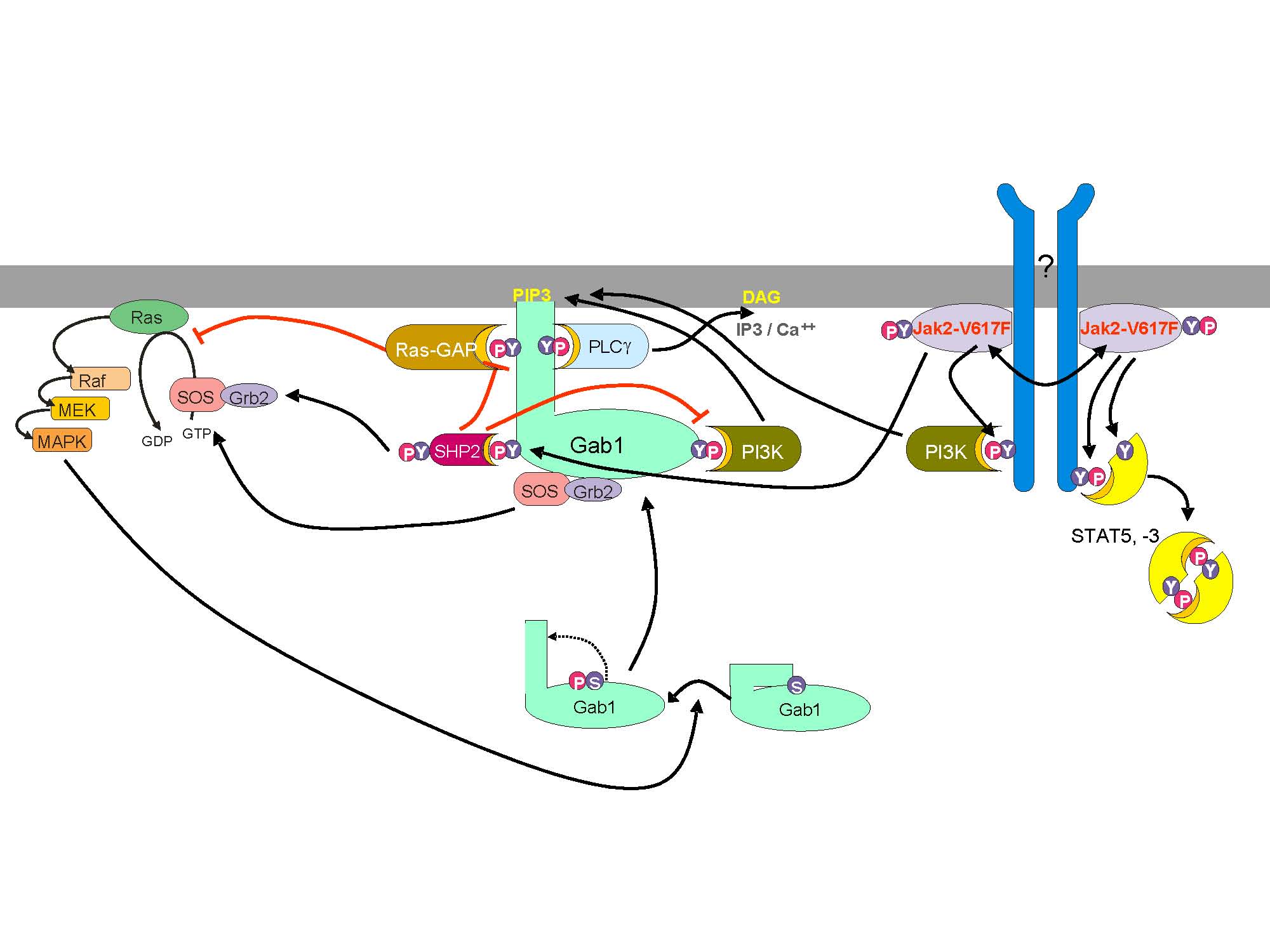
The interdisciplinary research project JAK-Sys addresses the understanding of the complex signalling and influencing factors as well as the identification of therapeutic targets for myeloproliferative neoplasm (MPN). MPNs are a group of diseases of the bone marrow, in which most of the haematopoiesis occurs. The activating JAK2-V617F mutation is found in the majority of myeloid disorders, which constitutes that it and its specific signalling pathways are attractive therapeutic targets. On the other hand JAK2 signalling is not only a factor for neoplastic cells, but also necessary for the normal haematopoiesis. Hence the complete eradication of the malignant clone is no therapeutic option. |
|
Interleukin-6 classic- and trans-signaling and therapeutic treatments
|
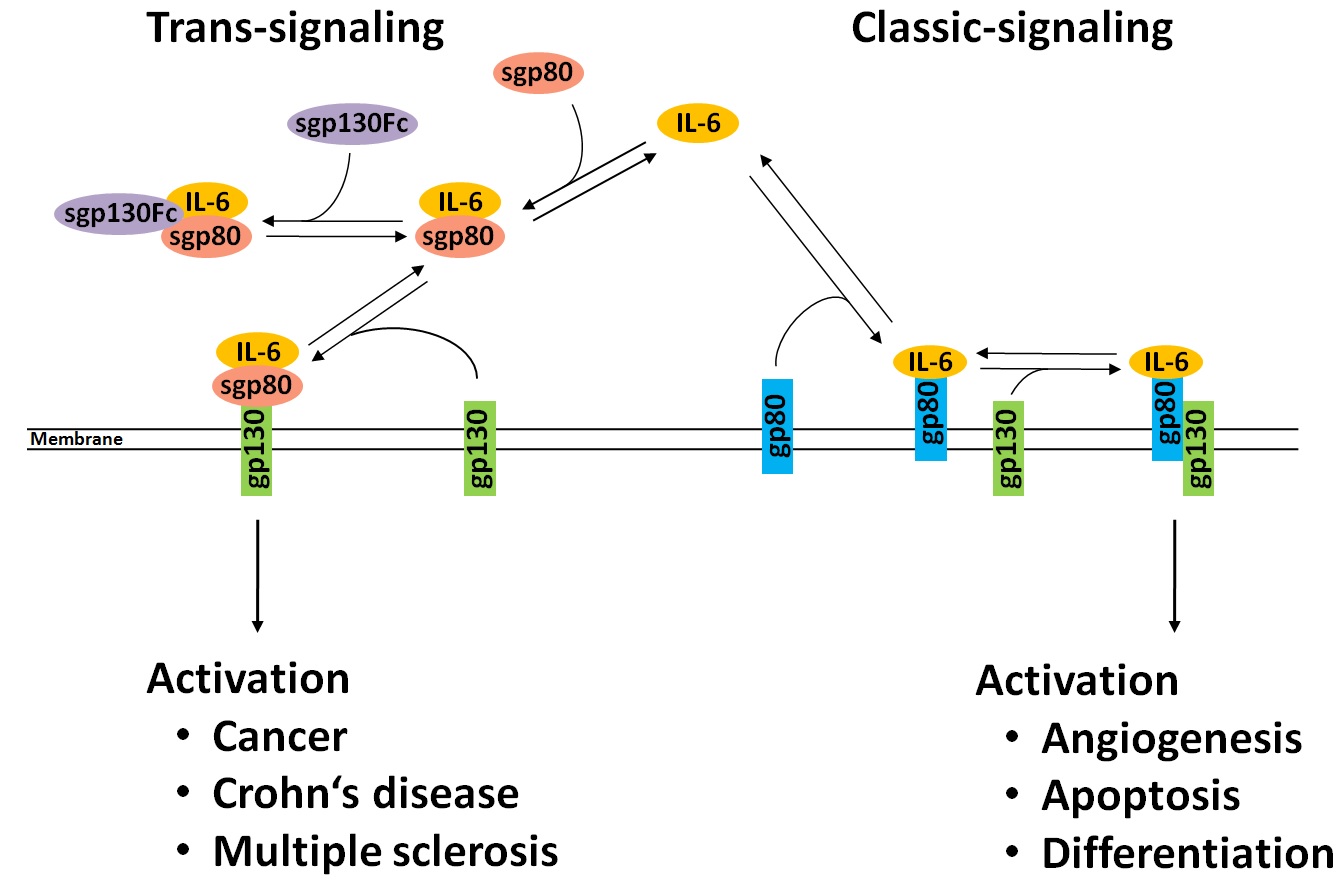
Interleukin-6 (IL-6) is a multipotent cytokine that is involved in many important cellular functions, however dysregulation of IL-6 production may induce numerous pathological states and was correlated to tumor proliferation.
It is known that the IL-6 signal is processed by two different pathways. Classic-signaling is induced by binding of IL-6 to a IL-6 receptor complex containing a membrane-bound subunit glycoprotein 80 (gp80). In contrast the involvement of soluble gp80 (sgp80) induces trans-signaling, which was connected to harmful diseases. An anti-inflammatory designer protein (sgp130Fc), which was developed to inhibit trans-signalling, potentially also inhibits classic-signalling. |
|
Guaranteed estimation, classification, and analysis of biochemical
and gene-regulation networks
|
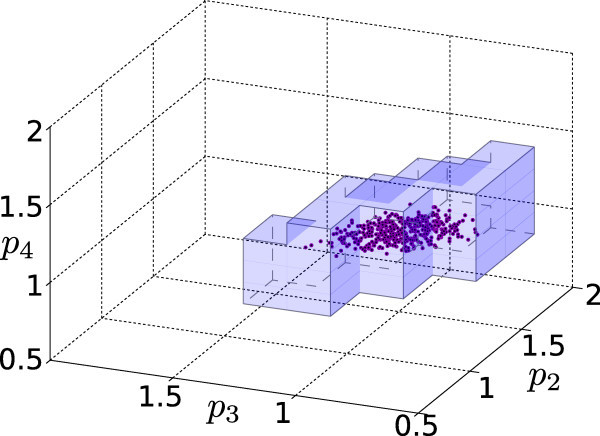
Biological and cellular processes can typically be represented as polynomial mathematical models, whose
analysis provides a complement to elaborative experimental studies. Obtaining reliable quantitative models however
is often very difficult, as available structural and quantitative knowledge is frequently limited; model
parameters can very frequently not be determined directly, and thus have to be estimated from uncertain measurements.
|
|
JAK-STAT and MAPK cascades
|
Signal transduction pathways are of essential importance for organisms to react to environmental changes, e.g. by regulating gene expression. One possible stimulus are cytokines that are involved in many inflammatory diseases. An important cytokine is Interleukin-6 (IL-6). Activation of the IL-6 signalling pathways lead to the initiation of the JAK/STAT pathway (Janus Kinase/Signal Transducer and Activator of Transcription) as well as the MAPK cascade (Mitogen Activated Protein Kinase). A deregulation of the described processes causes a variety of disease, e.g. rheumatoid arthritis, multiple sclerosis, disorders of the bone marrow and several types of tumors [9,10,11].
|

|
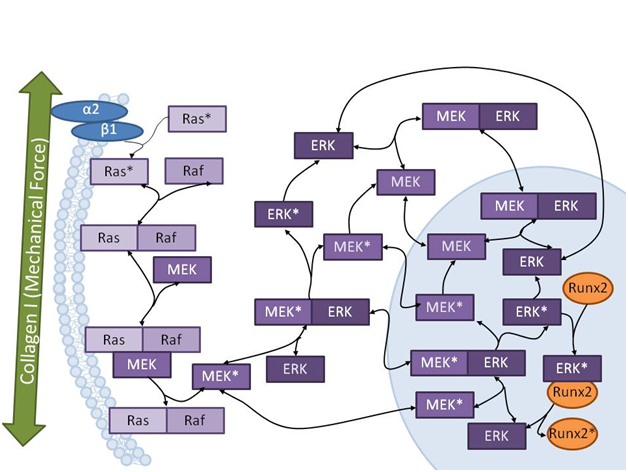
Modeling and control of mesenchymal stem cells
|
The project addresses the development and integration of new experimental and theoretical tools to elucidate and consequently predict quantitatively mechanisms of adult stem cell differentiation subject to mechanical, biochemical and physical stimuli of the matrix. The ultimate aim is to apply the generated knowledge and established tools for tissue engineering of human mesenchymal stem cells (MSC) as a source for cartilage and bone replacement in regenerative medicine.
|
|
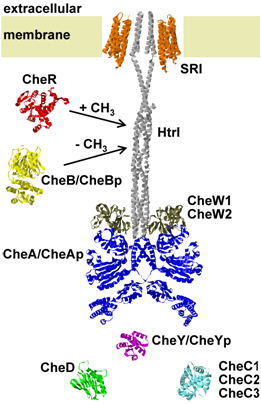
Phototaxis
|
Phototaxis is the swimming response to light and it allows a cell to find the best places (in terms of light conditions) in its environment. The phototaxis signal transduction network of Halobacterium salinarum has been intensively experimentally investigated [4,5,8] and predictive mathematical models have been developed [8]. However, still many molecular mechanisms are not understood and are therefore not included in the mathematical models.
|
|
Mathematical Modeling and Analysis of Bone Adaptation
|
Bone is a living tissue constantly adapting its shape
and microstructure to suit the changing biophysical environment.
To achieve this adaption ability, bone surfaces need to be destroyed and rebuilt immediately afterwards,
going through a tissue mechanism known as bone remodeling.
|
|
Guaranteed estimation and analysis for bioprocess engineering
|
Genetically engineered cells are nowaday used for the production of specified enzymes
requiered e.g. for pharmaceutical and environmental purposes. This project aims to analyse and enhance
the growth conditions of (human) cells in batch and continous processing.
Cell growth and production depends on the availability of substrates as well as on by-products (inhibiting effects).
|
|
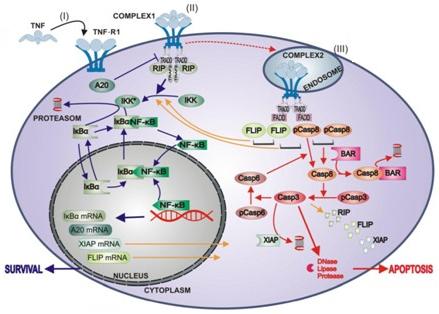
Tumor Necrosis Factor (TNF)-induced apoptosis: experiments,
modelling and model analysis
|
The pro-inflammatory cytokine TNF simultaneously induces survival pathways via activation of the nuclear factor κB, and apoptotic signaling by caspase activation when activating its cognate membrane receptor, TNF receptor type 1. We are studying the differential dose-response relationships of these two major TNF signaling pathways using a quantitative and dynamic modelling approach to investigate the crosstalk mechanisms of pro- and anti-apoptotic signaling as a function of dosage and time, and leading to the final outcome of the cellular response. |
|
Model-aided microaerobic process control
|
The goal of this project is to maximize productivity of photosynthetic
membranes in R. rubrum under dark, microaerophilic conditions. For
this reason, a growth model of R. rubrum in such conditions is
constructed utilizing the set-based approaches developed by the group.
|
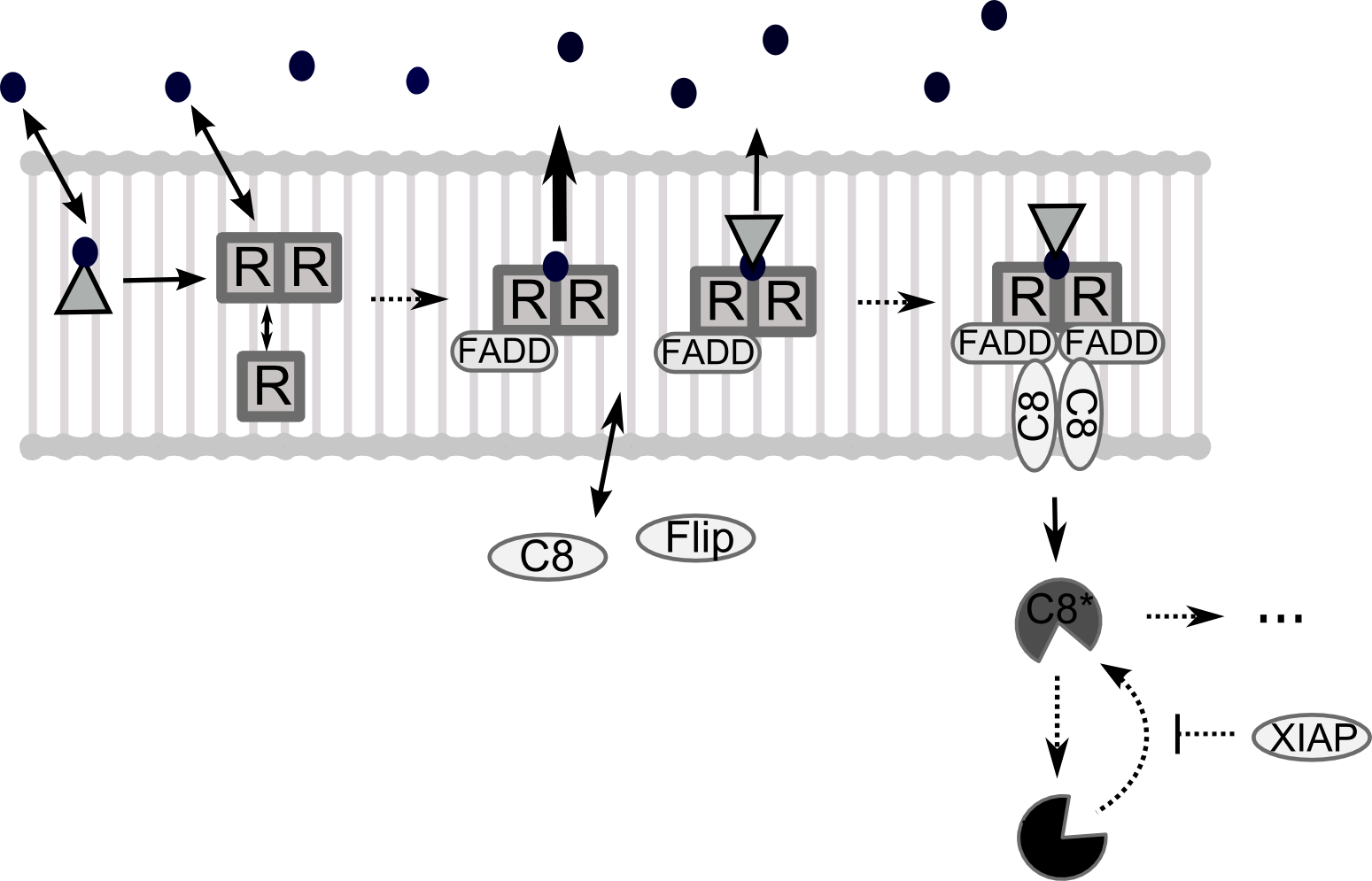
Trail-induced apoptosis toward predictive cancer therapy
|
Trail is a cytokine known to induce apoptosis (controlled cell death) in tumoral cells with little side-effects on healthy cells.
Furthermore, modifications of the Trail molecule (Fusionproteins) enhance the desired effects on cancer cells.
|
|
Journal Articles (peer reviewed)
S. Blacher, C. Erpicum, B. Lenoir, J. Paupert, G. Moraes, S. Ormenese, E. Bullinger, & A. Noël (2014). Cell invasion in the spheroid sprouting assay: A spatial organisation analysis adaptable to cell behaviour. PLoS ONE, 9(5):e97019.
M. C. Readman, M. Schliemann, D. Kalamatianos, & E. Bullinger (2013). A feedback control perspective on models of apoptosis signal transduction. Chaos, Solitons & Fractals, 50:93-99. Special Issue ‘Functionality and Dynamics in Biological Systems’.
P. Rumschinski, S. Streif, & R. Findeisen (2012). Combining qualitative information and semi-quantitative data for guaranteed invalidation of biochemical network models. Int. J. Robust Nonlin. Control, 22(10):1157-1173.
S. Streif, A. Savchenko, P. Rumschinski, S. Borchers, & R. Findeisen (2012). ADMIT: a toolbox for guaranteed model invalidation, estimation and qualitative-quantitative modeling. Bioinformatics, 28(9):1290-1291.
S. Borchers, S. Bosio, R. Findeisen, U. Haus, P. Rumschinski, & R. Weismantel (2011). Graph problems arising from parameter identification of discrete dynamical systems. Mathematical Methods of Operations Research, 73(3):381-400.
M. Schliemann, E. Bullinger, S. Borchers, F. Allgöwer, R. Findeisen, & P. Scheurich (2011). Heterogeneity reduces sensitivity of cell death for TNF-stimuli. BMC Syst Biol, 5:204.
J. Hasenauer, P. Rumschinski, S. Waldherr, S. Borchers, F. Allgöwer, & R. Findeisen (2010). Guaranteed steady state bounds for uncertain (bio-)chemical processes using infeasibility certificates. J. Proc. Contr., 20(9):1076-1083.
P. Rumschinski, S. Borchers, S. Bosio, R. Weismantel, & R. Findeisen (2010). Set-based dynamical parameter estimation and model invalidation for biochemical reaction networks. BMC Syst Biol, 4:69.
S. Streif, D. Oesterhelt, & W. Marwan (2010). A predictive computational model of the kinetic mechanism of stimulus-induced transducer methylation and feedback regulation through CheY in archaeal phototaxis and chemotaxis, BMC Syst Biol, 4(27)
S. Streif, S. Waldherr, F. Allgöwer, & R. Findeisen (2009). Steady state sensitivity analysis of biochemical reaction networks: A brief review and new methods. In A. Jayaraman and J. Hahn, editors, Systems Analysis of Biological Networks, Methods in Bioengineering, pages 129-148. Artech House MIT Press.
S. Streif, W. F. Staudinger, D. Oesterhelt, & W. Marwan (2009). Quantitative analysis of signal transduction in motile and phototactic cells by computerized light stimulation and model based tracking. Rev Sci Instrum, 80(2):023709.
S. Streif, W. F. Staudinger, W. Marwan, & D. Oesterhelt (2008). Flagellar rotation in the archaeon Halobacterium salinarum depends on ATP. J Mol Biol, 384:1-8.
S. Maldonado, R. Findeisen & F. Allgöwer (2008). Describing Force Induced Bone Growth and Adaptation by a Mathematical Model. J Musculoskeletal and Neuronal Interactions, 8(1):15--17.
S. Maldonado, R. Findeisen & F. Allgöwer (2008). Understanding the process of force-induced bone growth and adaptation through a mathematical model. Bone, 42(1): S61.
Conference Articles (peer reviewed)
N. Rudolph, T. Meyer, K. Franzen, C. Garbers, S. Streif, F. Schaper, A. Dittrich and R. Findeisen. A Two-level Approach for Fusing Early Signaling Events and Long Term Biological Responses. In International Symposium on Advanced Control of Chemical Processes (ADCHEM), pages 1229-1234, Whistler, Canada, June 2015.
S. Streif, N. Strobel and R. Findeisen. Inner Approximations of Consistent Parameter Sets Via Constraint Inversion and Mixed-integer Linear Programming. In Proc. of the 12th IFAC Symposium on Computer Applications in Biotechnology (CAB), pages 326-331, Mumbai, India, December 2013.
M. Schliemann, S. Livingstone, M. C. Readman, D. Kalamatianos, & E. Bullinger (2013). Quantifying heterogeneity of cell death. In Proc. of the 12th IFAC Symposium on Computer Applications in Biotechnology, Mumbai, India, 16-18 December 2013, pp. 181-186.
M. Schliemann, R. Findeisen, & E. Bullinger (2012). Robustness-based model validation of an apoptosis signalling network model. In Proc. of the 16th IFAC Symposium on System Identification, Brussels, Belgium, 11-13 July 2012, pages 930-935.
S. Borchers, P. Rumschinski, S. Bosio, R. Weismantel, & R. Findeisen (2009). A set-based framework for coherent model invalidation and parameter estimation of discrete time nonlinear systems. In Proc. of the 48th IEEE Conf. on Decision and Control, pages 6786-6792, Shanghai, China.
S. Maldonado, F. Allgöwer & R. Findeisen (2009). Global Sensitivity Analysis of Force-induced Bone Growth and Adaptation using Semidefinite Programming. In Proc. of the Foundations of Systems Biology and Engineering Conference (FOSBE), pages 141--144, Denver, USA.
Geffen, D., R. Findeisen, M. Schliemann, F. Allgöwer & M. Guay (2008). Observability Based Parameter Identifiability for Biochemical Reaction Networks. American Control Conference, Seattle, WA, USA, 11-13 June 2008, pp. 2130-2135.
Geffen, D., R. Findeisen, M. Schliemann, F. Allgöwer & M. Guay (2007). The question of parameter identifiability for biochemical reaction networks considering the NF-κB signal transduction pathway. 2nd Foundations of Systems Biology in Engineering FOSBE 2007, 9-12 September 2007, pp. 545-552.
Schliemann, M., T. Eißing, P. Scheurich & E. Bullinger (2007). Mathematical modelling of TNF-α induced apoptotic and anti-apoptotic signalling pathways in mammalian cells based on dynamic and quantitative experiments. 2nd Foundations of Systems Biology in Engineering FOSBE 2007, Stuttgart, Germany, 9-12 September 2007, pp. 213-218.